Effects of HLA Typing Resolution on Hardy-Weinberg Equilibrium, Linkage Disequilibrium, and Hidden Heterozygosity
Abstract
Objective:
The resolution level of human leukocyte antigen (HLA) typing critically influences the interpretation of population genetic parameters. This study aimed to quantitatively evaluate the effects of 2-, 4-, and 8-digit typing resolutions on allelic diversity, Hardy-Weinberg equilibrium (HWE), linkage disequilibrium (LD), and asymmetric LD (ALD) in a Central Anatolian population.
Materials and Methods:
High-resolution next-generation sequencing (NGS)-based HLA typing was performed for six loci (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1) in 150 unrelated healthy donors. Population genetic analyses were conducted using PyPop v1.2.1 software, and the results were comparatively assessed across 2-, 4-, and 8-digit resolution levels.
Results:
Higher typing resolution systematically increased allelic richness and revealed hidden heterozygosity. While several loci appeared to conform to HWE at lower resolutions, significant deviations emerged at higher resolutions. Linkage disequilibrium strength increased with resolution, and ALD analysis consistently demonstrated directional dominance between specific locus pairs, reflecting biological hierarchies within haplotypes. Low-resolution typing underestimated allelic diversity, obscured heterozygosity, masked evolutionary signals, and weakened LD, potentially leading to misinterpretations.
Conclusion:
High-resolution HLA typing, particularly at the 8-digit level, is essential for the accurate interpretation of population genetic parameters, disease association studies, and donor–recipient matching in transplantation. Future studies should focus on developing cost-effective high-resolution typing methods to enhance global accessibility and improve data accuracy in global HLA research.
Keywords:
HLA population, genetics next-generation, sequencing typing, resolution Hardy-Weinberg, equilibrium linkage, disequilibriumIntroduction
The human leukocyte antigen (HLA) complex, encoded by the major histocompatibility complex (MHC), is characterized by its remarkable polymorphism and linkage disequilibrium (LD), which makes it crucial for both immunity and disease susceptibility (1). Balancing selection, which promotes a heterozygote advantage and enhances resistance to infection, is the main evolutionary force maintaining this diversity (2). This diversity poses a significant challenge in clinical settings, such as hematopoietic stem cell transplantation, whereas strong LD aids in forming ancestral haplotypes that are essential for tracking population migration and disease susceptibility (3).
Identifying HLA alleles depends on typing resolution, which ranges from 2-digit serological groups to 8-digit sequences that capture the most detailed level of allele diversity. Population genetics utilizes tools such as the Hardy-Weinberg equilibrium (HWE) and LD to analyze a population's genetic structure (1-3). However, the quantitative effect of the "resolution gap" on these parameters remains unclear. Low-resolution methods can group functionally distinct alleles, leading to a "dilution effect", which can obscure true genetic diversity and weaken statistical signals (4,5).
This study addressed a key methodological question: How does HLA typing resolution influence the estimation of population genetic parameters and the evolutionary inferences drawn from them? Specifically, our objectives were to:
Quantify changes in allelic diversity and heterozygosity at 2-, 4-, and 8-digit resolutions.
Assess the effect of resolution on the results of HWE and Ewens-Watterson (EW) neutrality tests; and
Evaluate how typing resolution alters the strength and directionality of LD and asymmetric LD (ALD).
By analyzing a single Central Anatolian population at three distinct resolution levels, we aimed to demonstrate that typing resolution is a fundamental determinant of how population genetic structure and evolutionary patterns are interpreted (3,6,7).
Materials and Methods
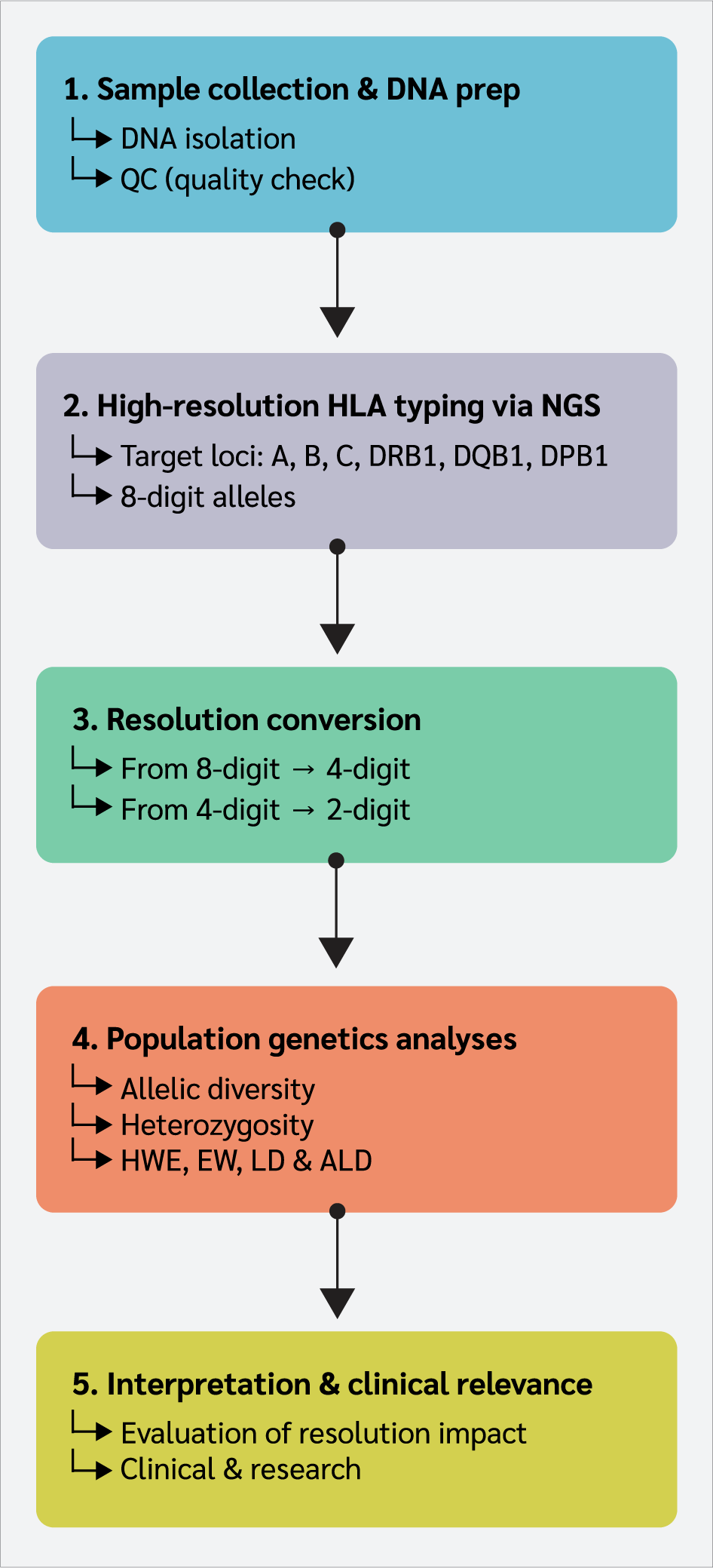
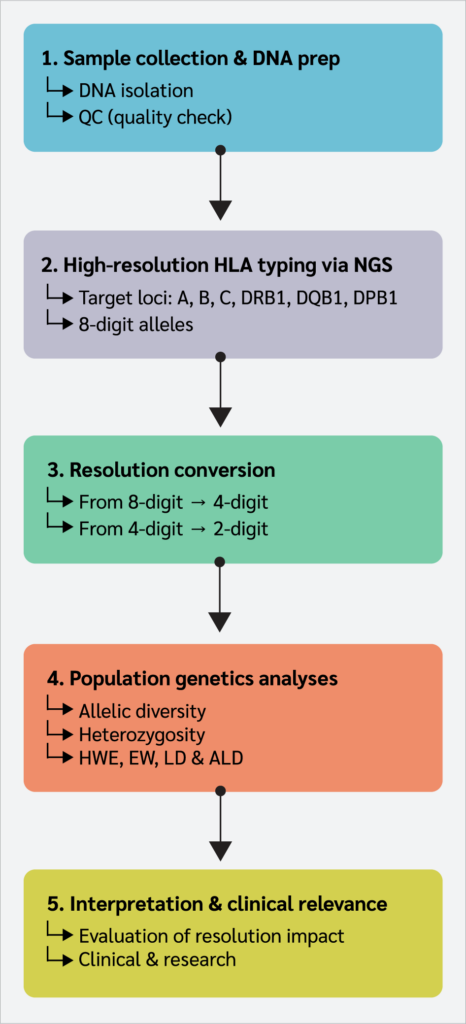
The overall workflow of the study is summarized in Figure 1.
Study Population and Methodological Considerations
This retrospective study included 150 unrelated healthy donors from the database of the Tissue Typing Laboratory, Eskişehir Osmangazi University, between January 2020 and July 2025. The study group consisted of 150 unrelated donors (52.7% female, 47.3% male) with a mean age of 38.6 ± 19.4 years. The cohort partially overlapped with the donor database used in our previous publication, while also incorporating newly recruited donors (8).
Most participants were of Central Anatolian origin, residing primarily in the city where the study was conducted or in neighboring provinces (8). As this was a retrospective study using a donor database, certain limitations were acknowledged. To minimize bias, individuals with known familial relationships were excluded based on a database cross-check and self-reporting by donors, ensuring that all participants were unrelated. No additional confounding factors were controlled for, as the primary objective was to compare the methodological resolutions within the same cohort rather than to conduct a case-control analysis.
The study was conducted in accordance with the principles of the Declaration of Helsinki, and the study protocol was approved by the Ethics Committee of Eskisehir Osmangazi University Faculty of Medicine (Approval No: 2025-06).
Human Leukocyte Antigen Typing
Genomic DNA was isolated from peripheral blood collected into tubes with K3 EDTA using an automated system (EZ1 DNA Blood Kit 200 µL; Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA concentrations were measured using the QIAxpert System (QIAGEN GmbH, Hilden, Germany), as described in our previous study (8,9). DNA samples purified to an A260/A280 ratio between 1.65 and 1.80 were used for subsequent next-generation sequencing (NGS) analysis.
High-resolution genotyping for HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 loci was conducted using the MIA FORA NGS FLEX HLA Typing Kit (BioArray Solutions Ltd., New Jersey, USA), following the manufacturer's guidelines. Sequencing data were processed using the MIA FORA NGS FLEX HLA Genotyping Software (version 3.0), with reference to the IMGT/HLA database version 3.43.0 (10). For comparative analyses, 8-digit high-resolution data were converted to 2- and 4-digit resolutions by focusing on the initial two fields of the HLA allele identifiers.
Population Genetics Analysis
Population genetics analyses for all three resolution levels were performed using Python for Population Genomics (PyPop) v1.2.1 software (11). Alleles with frequencies below 1% were excluded from the HWE and LD analyses to minimize the potential for statistical artifacts arising from rare variants.
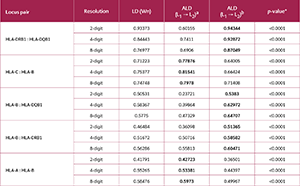
Compliance with HWE was evaluated using the Markov chain Monte Carlo (MCMC) exact test (12). Evidence of natural selection pressure was assessed via Slatkin's EW neutrality test (13). LD was measured using the normalized D′ coefficient and Wn (a multiallelic extension of the r² correlation measure) (14), and ALD analysis was performed based on the criteria defined by Thomson and Single (15). The statistical significance of LD measures was determined by a permutation test (1000 permutations) to account for multiple pairwise comparisons. For all analyses, statistical significance was set at p<0.05.
Results
Impact of Resolution on Allelic Diversity and Genotype Counts
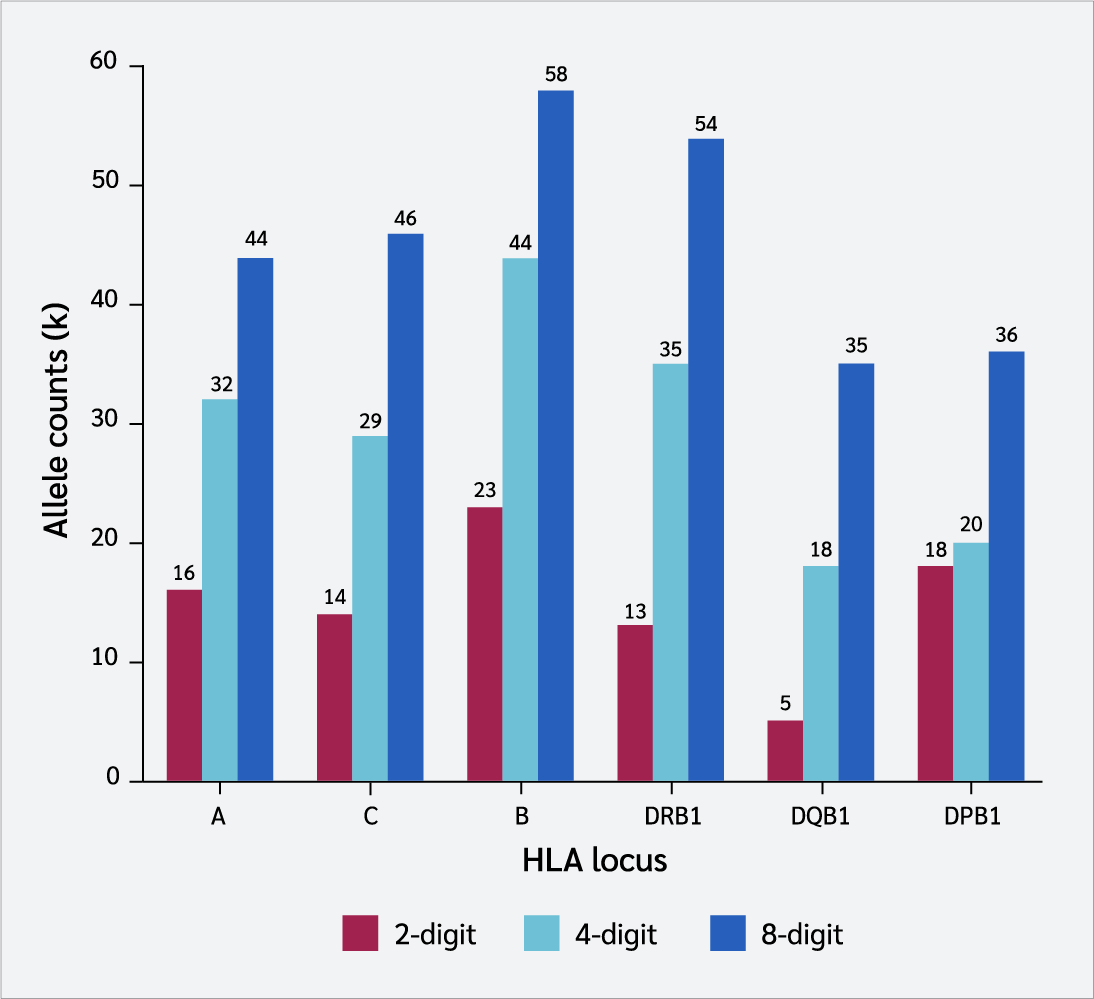
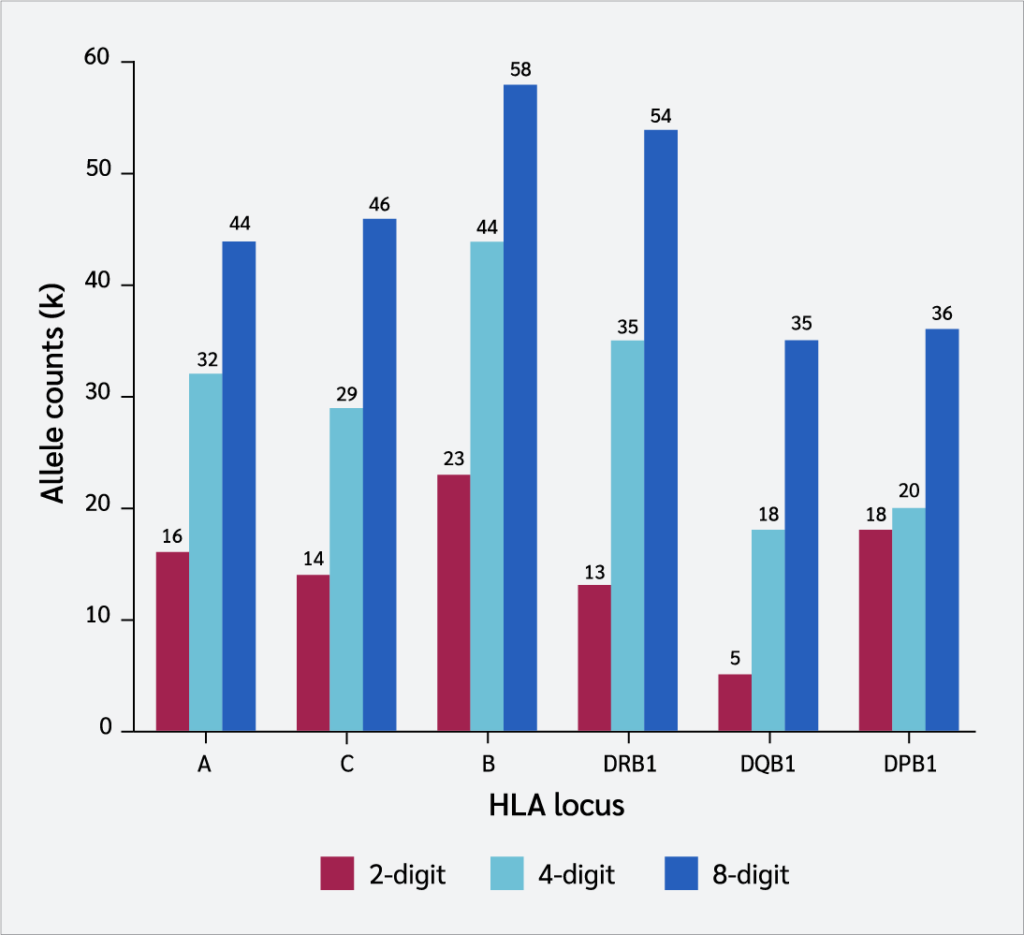
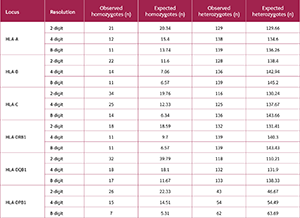
Analysis of six HLA loci demonstrated that as typing resolution improved, the number of distinct alleles (k) also increased (Figure 2). For instance, transitioning from a 2-digit to an 8-digit resolution increased allele richness from 16 to 44 for the HLA-A locus and from 23 to 58 for HLA-B.
Alongside the increase in allele numbers, there was a consistent decrease in the number of homozygous individuals across all loci with higher resolution, whereas the number of heterozygous individuals increased (Table 1). For example, at the HLA-C locus, the number of observed homozygotes dropped from 34 at a 2-digit resolution to 14 at an 8-digit resolution. By contrast, the number of heterozygous individuals at this locus increased from 116 to 136.
Hardy-Weinberg Equilibrium and Natural Selection Test Results
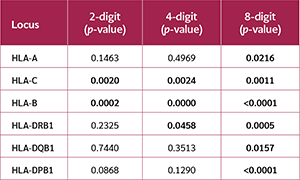
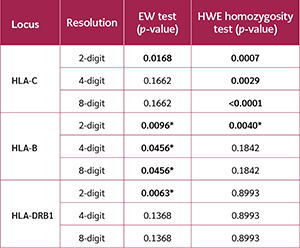
The population's adherence to the HWE was highly sensitive to typing resolution (Table 2). The HLA-C and HLA-B loci consistently and significantly deviated from HWE across all three resolution levels (all p<0.01). In contrast, the HLA-A, HLA-DQB1, and HLA-DPB1 loci conformed to HWE at lower resolutions but deviated significantly at the 8-digit level. The HLA-DRB1 locus deviated at both 4-digit (p=0.0458) and 8-digit (p=0.0005) resolutions.
The EW neutrality test revealed a different and more complex pattern (Table 3). The HLA-B locus showed significant deviation from neutrality at all resolutions. However, the HLA-C, HLA-DRB1, and HLA-DQB1 loci deviated significantly only at the 2-digit level, whereas the HLA-A and HLA-DPB1 loci did not deviate at any resolution.
Linkage Disequilibrium (LD) and Asymmetric LD (ALD)
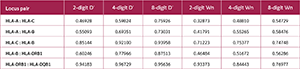
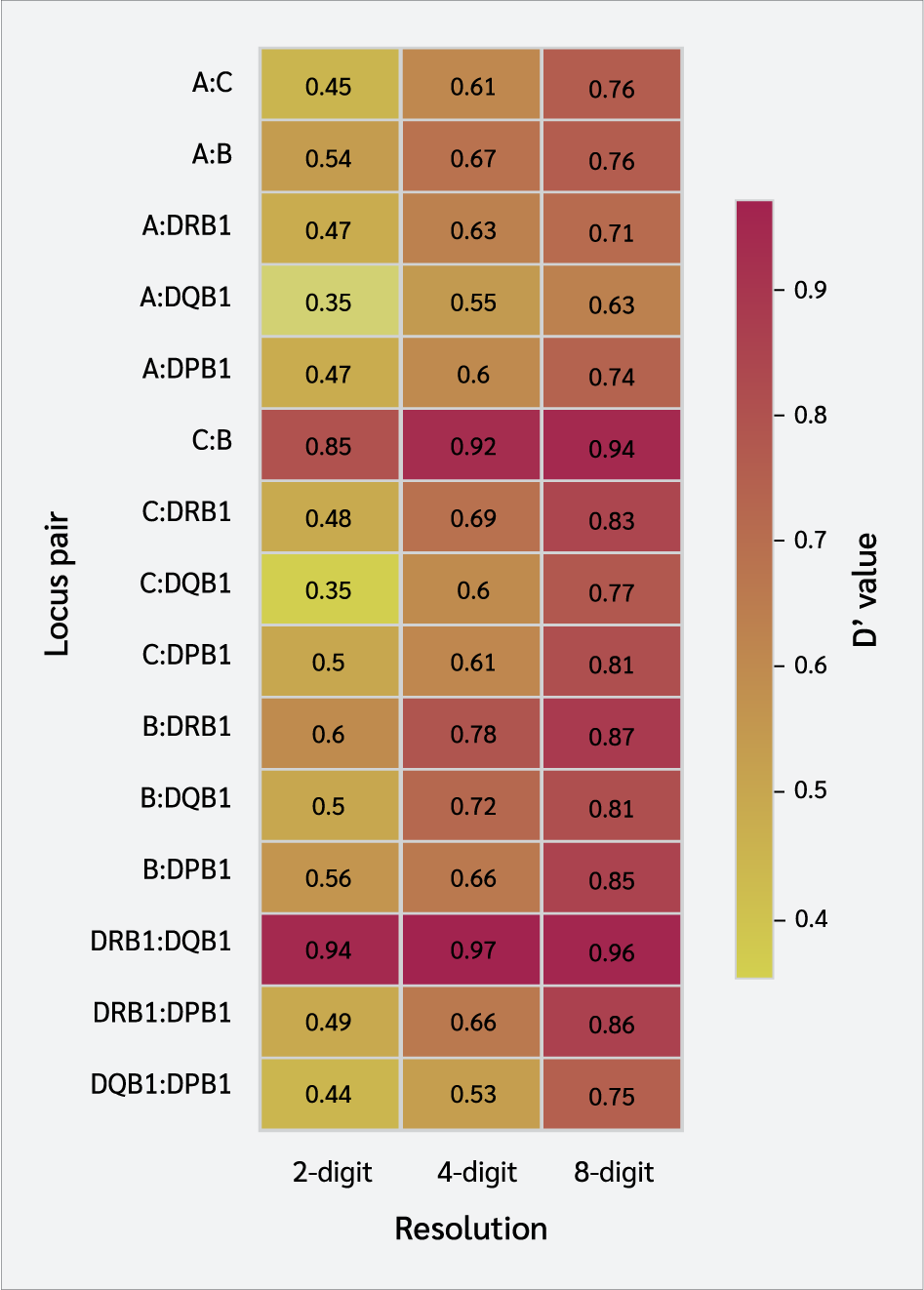
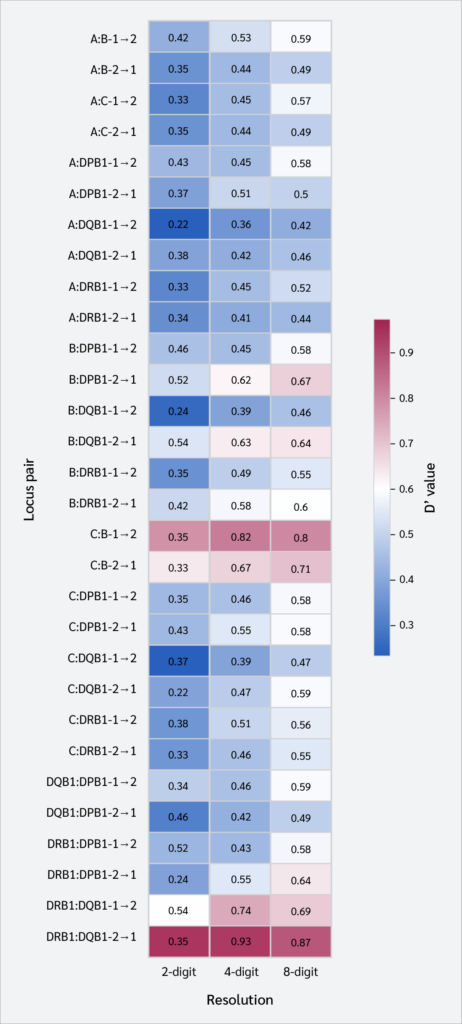
The normalized D′ coefficient was used to assess the LD between pairs of loci. Generally, as typing resolution increased, there was a noticeable increase in the D′ values (Table 4, Figure 3, Supplementary Table 1). For instance, the D′ value for the HLA-A : HLA-C locus pair was 0.46928 at the 2-digit level and increased to 0.75926 at the 8-digit level. The strongest LD signals were detected at the 8-digit level between HLA-C : HLA-B (D′=0.93958) and HLA-DRB1 : HLA-DQB1 (D′=0.95636).
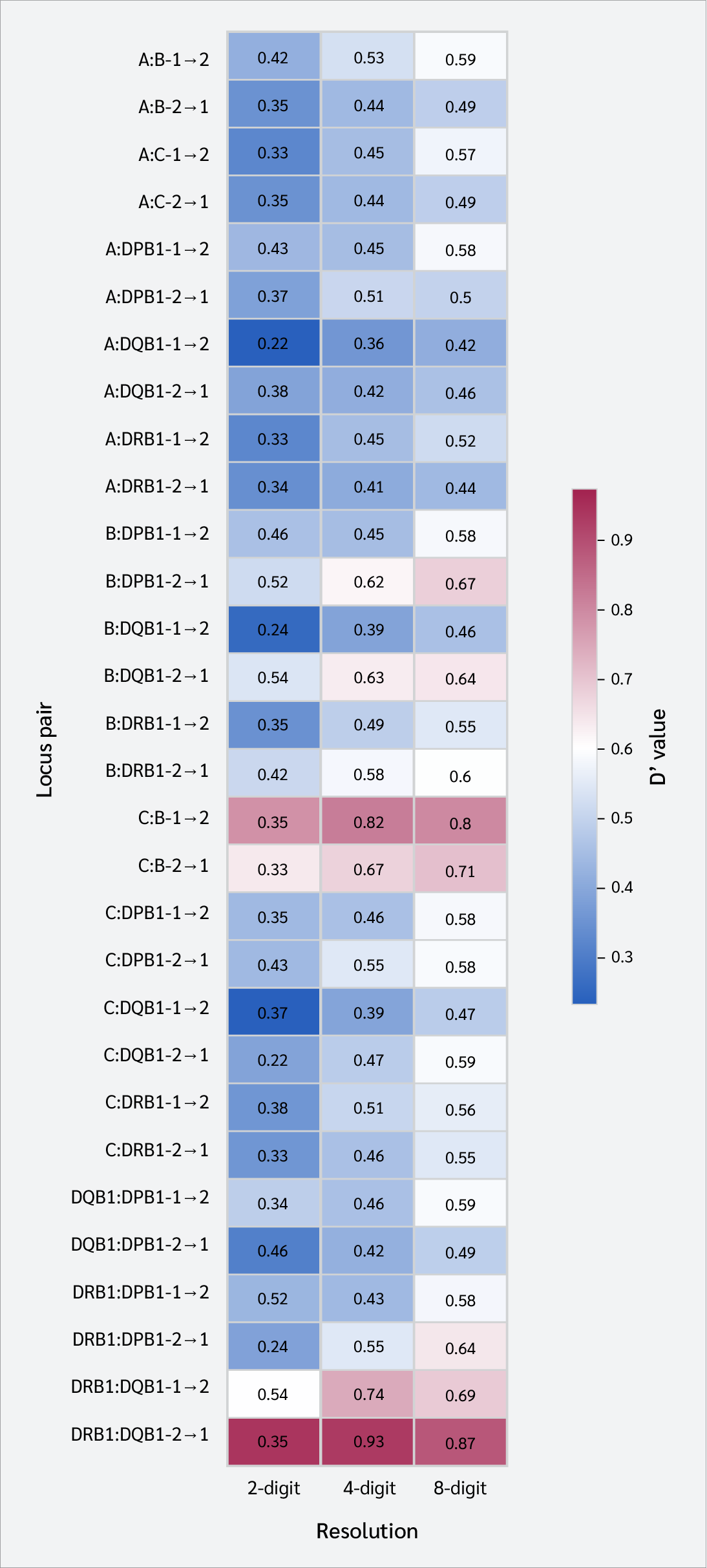
In the ALD analysis, a consistent and prevailing direction was observed between certain locus pairs, regardless of resolution levels (Table 5, Figure 4, Supplementary Table 2). Across all resolution levels analyzed, the predictive influence from HLA-C to HLA-B (C → B) and from HLA-DQB1 to HLA-DRB1 (DQB1 → DRB1) was more pronounced than that in the reverse direction.
Discussion
The findings of this study demonstrated that increasing HLA typing resolution, from low to high, reshapes our understanding of the HLA genetic landscape. High-resolution typing uncovered previously "hidden" allelic diversity and heterozygosity, prompting a re-evaluation of HWE, natural selection, and LD. Our results highlighted how the "dilution effect" associated with low-resolution typing systematically diminishes or obscures evolutionary signals, resulting in a "blurred" view of the genetic landscape (16).
"Dilution Effect": The Core Mechanism Obscuring the Genetic Landscape
The primary mechanism responsible for nearly all observed outcomes is the "dilution effect," which stems from the low resolution (8). Two-digit typing merges structurally and functionally distinct alleles (e.g., C∗07:01 and C∗07:02) into a single serological category (C*07). This leads to an artificial decrease in the number of alleles, as demonstrated in our study: the number of alleles identified at the HLA-A locus increased from 16 at 2-digit to 32 at 4-digit and 44 at 8-digit resolution.
More critically, this merging leads to the artificial classification of individuals with different genotypes (e.g., C*07:01/C*07:02) as "homozygous" (C*07/C*07). This phenomenon, termed "hidden heterozygosity," directly accounts for the systematic reduction in the number of homozygotes observed in our study as resolution improved (11).
Selection at the Haplotype Level: Synthesis of Complex Signals in HWE and EW Tests
Our analysis also demonstrated that deviations from the HWE are notably influenced by resolution and become particularly complex when considered in conjunction with selection tests. The HLA-C and HLA-B loci consistently deviated from HWE across all resolution levels (all p≤0.0024), indicating the strong selection pressure. However, selection signals at these loci are not well understood. Hardy-Weinberg equilibrium analyses revealed that the number of homozygous individuals at these loci was significantly higher than expected. In contrast, the EW neutrality test indicated that the overall homozygosity (F) at these loci was lower than expected under neutral evolution, suggesting a heterozygote advantage.
This apparent contradiction may result from selection acting on the entire haplotype rather than on the individual alleles. If a beneficial B-C haplotype becomes common in the population, certain genotypes may appear overrepresented in HWE analyses (17). This was more apparent when examining specific targets of selection at the genotypic level. The overall excess of homozygotes at the HLA-C locus was specifically driven by genotypes, such as C*07+C*07. The presence of many other rare haplotypes in the population maintained a low overall homozygosity (F), creating a heterozygote advantage signal in the EW test (18). Thus, the differing outcomes of the two tests revealed two distinct aspects of the same evolutionary process: the rise of certain advantageous haplotype blocks and the maintenance of overall diversity (19).
Linkage Disequilibrium and the True Power of Ancestral Haplotypes
Strong LD, the fundamental architecture of the HLA region, became increasingly evident with higher typing resolution in our study. This is a direct consequence of the elimination of the "dilution effect.” High-resolution analysis directly reflected the true, non-random association between specific alleles and leads to a marked increase in LD measures such as D'. For example, the D′ value for the HLA-A : HLA-C locus pair increased from 0.469 at the 2-digit level to 0.759 at the 8-digit level. This increase was particularly notable for some pairs; for instance, at 8-digit resolution, the D' value for the HLA-B : HLA-DRB1 pair was over 12% higher than that observed at 4-digit resolution.
These findings suggest that high-resolution typing does not create new links, but rather reveals the inherent strength of ancestral haplotype structures that have been maintained by evolutionary forces. In particular, the HLA-C : HLA-B pair in Class I and the HLA-DRB1 : HLA-DQB1 pair in Class II were identified as nearly stable genetic blocks, exhibiting exceptionally high LD values. This suggests that previous evolutionary research using low-resolution datasets may have consistently underestimated the extent of MHC haplotype conservation.
Asymmetric LD analysis further clarified these relationships. Although ALD values varied depending on typing resolution, directional dominance (e.g., C→B and DQB1→DRB1) remained consistent regardless of resolution in certain locus pairs. This can be seen as a biological reflection of the functional or evolutionary hierarchy within haplotypes (20).
Unveiling Hierarchical Structures in Haplotypes Through Asymmetric LD
Traditional LD metrics such as D' and r² offer a single, symmetric value to describe the relationship between two loci. However, in systems with multiple alleles, such as HLA, where one locus has significantly more alleles than the other, this method does not provide a complete picture. To address this issue, ALD was developed by assessing the correlation between loci in both directions.
A key finding of our study was that the directional dominance identified by ALD (e.g., DQB1 → DRB1 and C → B) remained consistent across all resolution levels, even with a substantial increase in the number of alleles. This consistency strongly indicates that the observed patterns are not merely statistical anomalies but reflect genuine biological and evolutionary processes.
Our results demonstrated that the W(DRB1/DQB1) value for the HLA-DRB1 : HLA-DQB1 pair was almost 1.00, indicating that knowing an allele at the HLA-DQB1 locus allows for the perfect prediction of the associated HLA-DRB1 allele. However, the predictive accuracy in the opposite direction, W(DQB1/DRB1), is not flawless. This asymmetry implies a functional or evolutionary hierarchy between these two loci. This situation might arise from selection pressures that aim to maintain the structural and functional integrity of the expressed DR-DQ heterodimer or from selection on one locus, causing a "genetic hitchhiking" effect on the other.
The Broad Implications and Study Limitations
The results of this study have significant implications for contemporary immunogenetic research. In the context of disease association studies, achieving high resolution is crucial for identifying the actual causal variant, as low-resolution findings can be deceptive (21). In the field of anthropological genetics, low resolution tends to systematically underestimate the strength of linkage disequilibrium, potentially leading to incorrect conclusions regarding population history. Regarding transplant compatibility, 8-digit resolution offers a vital evaluation of genomic compatibility throughout the entire linked MHC region, reducing the clinical risks associated with mismatches in minor histocompatibility antigens or immune-regulating genes, such as tumor necrosis factor (TNF) (22).
However, the broad adoption of high-resolution typing has encountered several obstacles. Increased expenses, the need for specialized equipment, and the demand for advanced bioinformatics expertise can pose significant challenges, especially in resource-constrained environments. Although this study established 8-digit typing as the scientific benchmark, future research should also aim to develop more affordable and accessible high-resolution methods to ensure that these advantages are globally applicable. Our findings, consistent with those of studies in other populations that reveal greater diversity with high-resolution, strongly support the investment in such technologies.
High-resolution HLA typing studies in African cohorts have demonstrated a marked increase in allelic richness and the unmasking of “hidden heterozygosity,” closely paralleling our observations in the Central Anatolian population (16). In contrast, lower-resolution analyses in European datasets (7) have tended to underestimate haplotype conservation, whereas our results confirm that higher resolution sharpens LD signals and more accurately reflects ancestral haplotype structures.
Moreover, methodological advances in asymmetric LD analysis (6) provided a theoretical framework for our findings on stable directional dominance (e.g., C→B, DQB1→DRB1) across all resolution levels. Clinically, studies on transplantation immunogenetics (22) have shown that low-resolution matching increases the risk of graft-versus-host disease and rejection. Our demonstration that 8-digit typing represents the gold standard is, therefore, both clinically and evolutionarily relevant.
Of note, theoretical models of heterozygote advantage (19) suggested that MHC diversity itself may drive the appearance of heterozygote advantage, offering a conceptual framework for the complex HWE and neutrality test outcomes that we observed. Taken together, these comparisons highlighted that our study not only provided a region-specific contribution but also reinforces broader international evidence that high-resolution typing is essential for accurate population genetic inference and clinical decision-making.
Conclusion
This study quantitatively demonstrated that the resolution of HLA typing had a significant influence on the interpretation of population genetic parameters. Low-resolution data can lead to inaccurate conclusions by underestimating allelic diversity, concealing heterozygosity, hiding evolutionary signals, and weakening the LD power. Our principal findings revealed that as the resolution improved, allelic diversity became more apparent, HWE results underwent critical changes, LD signals became more pronounced, and biological hierarchies were validated. Collectively, these findings establish that high-resolution typing, particularly at the 8-digit level, is the gold standard for enhancing HLA population genetics, improving disease association analyses, and optimizing donor-recipient matching in transplantation. Future research should focus on cost-effective, high-resolution typing approaches to enhance the global accessibility and applicability of these insights.
Ethical Approval
The study protocol was approved by the Non-Interventional Clinical Research Ethics Committee of Eskişehir Osmangazi University on April 29, 2025 with the decision number 2025-06.
Informed Consent
N.A.
Peer-review
Externally peer-reviewed
Author Contributions
Concept – E.Y.; Design – E.Y.; Supervision – E.Y.; Fundings – E.Y.; Materials – E.Y.; Data Collection and/or Processing – E.Y.; Analysis and/or Interpretation – E.Y.; Literature Review – E.Y.; Writer – E.Y.; Critical Reviews – E.Y.
Conflict of Interest
The author declares no conflict of interest.
Financial Disclosure
The author declared that this study has received no financial support.
Acknowledgment
The author gratefully acknowledges the staff of the Tissue Typing Laboratory, Eskişehir Osmangazi University Faculty of Medicine, for their assistance in the data collection process.
References
Radwan J, Babik W, Kaufman J, Lenz TL, Winternitz J. Advances in the evolutionary understanding of MHC polymorphism. Trends Genet. 2020;36(4):298-311. [CrossRef]
Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272(5258):67-74. [CrossRef]
Sakaue S, Gurajala S, Curtis M, Luo Y, Choi W, Ishigaki K, et al. Tutorial: a statistical genetics guide to identifying HLA alleles driving complex disease. Nat Protoc. 2023;18(9):2625-41. [CrossRef]
Listgarten J, Brumme Z, Kadie C, Xiaojiang G, Walker B, Carrington M, et al. Statistical resolution of ambiguous HLA typing data. PLoS Comput Biol. 2008;4(2):e1000016. [CrossRef]
Ewens WJ. The sampling theory of selectively neutral alleles. Theor Popul Biol. 1972;3(1):87-112. [CrossRef]
Single RM, Strayer N, Thomson G, Paunic V, Albrecht M, Maiers M. Asymmetric linkage disequilibrium: Tools for assessing multiallelic LD. Hum Immunol. 2016;77(3):288-94. [CrossRef]
Evseeva I, Nicodemus KK, Bonilla C, Tonks S, Bodmer WF. Linkage disequilibrium and age of HLA region SNPs in relation to classic HLA gene alleles within Europe. Eur J Hum Genet. 2010;18(8):924-32. [CrossRef]
Yantır E, Gündüz E, Çolak E. HLA alleles, genotype and haplotype analyzes from Central Anatolia Region of Turkey. Balkan Med J. 2023;40(5):358-66. [CrossRef]
Sahin Tekin M, Yorulmaz G, Yantir E, Gunduz E, Colak E. A novel finding of an HLA allele's and a haplotype's relationship with SARS-CoV-2 vaccine-associated subacute thyroiditis. Vaccines (Basel). 2022;10(12):1986. [CrossRef]
Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA database. Nucleic Acids Res. 2020;48(D1):D948-55. [CrossRef]
Lancaster AK, Nelson MP, Single R, Solberg O, Tsai Y, Meyer D, et al. PyPop: Python for population genomics. Version 1.2.1. Zenodo; 2025. [CrossRef]
Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48(2):361-72.
Slatkin M. An exact test for neutrality based on the Ewens sampling distribution. Genet Res. 1994;64(1):71-4. [CrossRef]
Lewontin RC. The interaction of selection and linkage. I. General Considerations; Heterotic Models. Genetics. 1964;49(1):49-67. [CrossRef]
Thomson G, Single RM. Conditional asymmetric linkage disequilibrium (ALD): extending the biallelic r2 measure. Genetics. 2014;198(1):321-31. [CrossRef]
Banjoko AW, Ng'uni T, Naidoo N, Ramsuran V, Hyrien O, Ndhlovu ZM. High resolution class I HLA-A, -B, and -C diversity in Eastern and Southern African populations. Sci Rep. 2025;15(1):23667. [CrossRef]
Meyer D, Thomson G. How selection shapes variation of the human major histocompatibility complex: a review. Ann Hum Genet. 2001;65(Pt 1):1-26. [CrossRef]
Slatkin M. Joint estimation of selection intensity and mutation rate under balancing selection with applications to HLA. Genetics. 2022;221(2):iyac058. [CrossRef]
Cherry JL. Heterozygote advantage cannot explain MHC diversity, but MHC diversity can explain heterozygote advantage. bioRxiv [Preprint]. May 31, 2025:2025.05.27.656382. [CrossRef]
Pedersen MB, Asmussen SR, Sarfelt FM, Saksager AB, Sackett PW, Nielsen M, et al. Integration of HLA-DR linkage disequilibrium to MHC class II predictions. bioRxiv [Preprint]. May 24, 2023: 2023.05.24.542040. [CrossRef]
Thorsby E. A short history of HLA. Tissue Antigens. 2009;74(2):101-16. [CrossRef]
Petersdorf EW. The major histocompatibility complex: a model for understanding graft-versus-host disease. Blood. 2013;122(11):1863-72. [CrossRef]
VOLUME
,
ISSUE
Correspondence
Received
Accepted
Published
Suggested Citation
DOI
License